VNeAT (Voxel-wise Neuroimaging Analysis Toolbox)
| Resource Type | Date |
|---|---|
| Software | 2017-06-29 |
Description
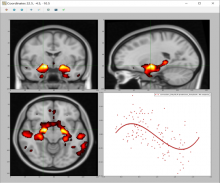
VNeAT (Voxel-wise Neuroimaging Analysis Toolbox) is a command-line toolbox written in Python that provides the tools to analyze the linear and nonlinear dynamics of a particular tissue and study the statistical significance of such dynamics at the voxel level.
Authors
| Name | Position / Role |
|---|---|
| Santi Puch Giner | Author |
| Asier Aduriz Berasategi | Author |
| Adrià Casamitjana Díaz | Contributor |
| Verónica Vilaplana Besler | Advisor (UPC) |
| Juan Domingo Gispert | Advisor (PMF)Institutions |
Institutions
Image and Video Processing Group, Universitat Politècnica de Catalunya
Pasqual Maragall Foundation
Publication
This work has been accepted at the NIPS 2016 Workshop on Machine Learning for Health.
The workshop paper is available on arXiv, and the associated poster is available here.
In order to cite this work please use the following BibTeX code:
@article{Puch_2016_NIPS4H,
author = {{Puch}, S. and {Aduriz}, A. and {Casamitjana}, A. and {Vilaplana}, V. and
{Petrone}, P. and {Operto}, G. and {Cacciaglia}, R. and {Skouras}, S. and
{Falcon}, C. and {Molinuevo}, J.~L. and {Domingo Gispert}, J.
},
title = "{Voxelwise nonlinear regression toolbox for neuroimage analysis: Application to aging and neurodegenerative disease modeling}",
journal = {ArXiv e-prints},
archivePrefix = "arXiv",
eprint = {1612.00667},
primaryClass = "stat.ML",
year = 2016,
month = dec
}
People involved
| Veronica Vilaplana | Associate Professor |
| Adrià Casamitjana | PhD Candidate |
Related Publications
|
, “Voxelwise nonlinear regression toolbox for neuroimage analysis: Application to aging and neurodegenerative disease modeling”, in NIPS 2016 Workshop on Machine Learning for Health, 2016. |
|
, “Analysis of the dynamics of gray matter reduction in Alzheimer's Disease”. 2016. |
|
, “Nonlinear analysis toolbox for neurodegenerative diseases and aging”. 2016. |
